| Chain sequence(s) | A: LDNGLARTPTMGWLHWERFMCNLDCQEEPDSCISEKLFMEMAELMVSEGWKDAGYEYLCIDDCWMAPQRDSEGRLQADPQRFPHGIRQLANYVHSKGLKLGIYADVGNKTCAGFPGSFGYYDIDAQTFADWGVDLLKFDGCYCDSLENLADGYKHMSLALNRTGRSIVYSCEWPLYMWPFQKPNYTEIRQYCNHWRNFADIDDSWKSIKSILDWTSFNQERIVDVAGPGGWNDPDMLVIGNFGLSWNQQVTQMALWAIMAAPLFMSNDLRHISPQAKALLQDKDVIAINQDPLGKQGYQLRQGDNFEVWERPLSGLAWAVAMINRQEIGGPRSYTIAVASLGKGVACNPACFITQLLPVKRKLGFYEWTSRLRSHINPTGTVLLQLENTM |
| Distance of aggregation | 10 Å |
| Dynamic mode | Yes |
The table below lists A3D score for protein residues. Residues with A3D score > 0.0000 are marked by yellow rows.
| residue index | residue name | chain | Aggrescan3D score | mutation |
|---|---|---|---|---|
| residue index | residue name | chain | Aggrescan3D score | |
| 32 | L | A | 1.1279 | |
| 33 | D | A | 0.0000 | |
| 34 | N | A | -1.3027 | |
| 35 | G | A | -0.9707 | |
| 36 | L | A | -0.4889 | |
| 37 | A | A | 0.0000 | |
| 38 | R | A | -1.4549 | |
| 39 | T | A | -0.9487 | |
| 40 | P | A | 0.0000 | |
| 41 | T | A | -0.5029 | |
| 42 | M | A | 0.0000 | |
| 43 | G | A | 0.0000 | |
| 44 | W | A | 0.0000 | |
| 45 | L | A | 0.0000 | |
| 46 | H | A | 0.0000 | |
| 47 | W | A | 0.0099 | |
| 48 | E | A | -1.3724 | |
| 49 | R | A | -0.1359 | |
| 50 | F | A | 0.8154 | |
| 51 | M | A | 0.0000 | |
| 52 | C | A | 0.0680 | |
| 53 | N | A | -0.5295 | |
| 54 | L | A | -0.0175 | |
| 55 | D | A | -1.9725 | |
| 56 | C | A | -2.0765 | |
| 57 | Q | A | -3.0582 | |
| 58 | E | A | -3.4991 | |
| 59 | E | A | -3.6705 | |
| 60 | P | A | -3.0501 | |
| 61 | D | A | -3.3723 | |
| 62 | S | A | 0.0000 | |
| 63 | C | A | 0.0000 | |
| 64 | I | A | 0.0000 | |
| 65 | S | A | -1.6992 | |
| 66 | E | A | -2.6330 | |
| 67 | K | A | -2.5529 | |
| 68 | L | A | 0.0000 | |
| 69 | F | A | 0.0000 | |
| 70 | M | A | -1.2209 | |
| 71 | E | A | -1.6103 | |
| 72 | M | A | 0.0000 | |
| 73 | A | A | 0.0000 | |
| 74 | E | A | -1.5936 | |
| 75 | L | A | -0.9118 | |
| 76 | M | A | 0.0000 | |
| 77 | V | A | -1.6161 | |
| 78 | S | A | -1.7613 | |
| 79 | E | A | -2.9444 | |
| 80 | G | A | -2.1989 | |
| 81 | W | A | 0.0000 | |
| 82 | K | A | -3.4667 | |
| 83 | D | A | -3.2050 | |
| 84 | A | A | -2.2730 | |
| 85 | G | A | -1.4764 | |
| 86 | Y | A | 0.0000 | |
| 87 | E | A | -1.4226 | |
| 88 | Y | A | -0.4737 | |
| 89 | L | A | 0.0000 | |
| 90 | C | A | 0.0000 | |
| 91 | I | A | 0.0000 | |
| 92 | D | A | 0.0000 | |
| 93 | D | A | -0.0279 | |
| 94 | C | A | 0.0000 | |
| 95 | W | A | 0.0000 | |
| 96 | M | A | 0.0000 | |
| 97 | A | A | 0.0000 | |
| 98 | P | A | -1.5843 | |
| 99 | Q | A | -2.8764 | |
| 100 | R | A | -4.0078 | |
| 101 | D | A | -3.8871 | |
| 102 | S | A | -2.6454 | |
| 103 | E | A | -3.3240 | |
| 104 | G | A | -3.1229 | |
| 105 | R | A | -3.5096 | |
| 106 | L | A | 0.0000 | |
| 107 | Q | A | -1.9351 | |
| 108 | A | A | -1.2757 | |
| 109 | D | A | -1.4199 | |
| 110 | P | A | -1.2061 | |
| 111 | Q | A | -2.0065 | |
| 112 | R | A | -2.3583 | |
| 113 | F | A | 0.0000 | |
| 114 | P | A | -1.9065 | |
| 115 | H | A | -1.8222 | |
| 116 | G | A | 0.0000 | |
| 117 | I | A | 0.0000 | |
| 118 | R | A | -2.8543 | |
| 119 | Q | A | -2.4216 | |
| 120 | L | A | 0.0000 | |
| 121 | A | A | 0.0000 | |
| 122 | N | A | -2.0112 | |
| 123 | Y | A | -0.4088 | |
| 124 | V | A | 0.0000 | |
| 125 | H | A | -1.7191 | |
| 126 | S | A | -1.0258 | |
| 127 | K | A | -1.4707 | |
| 128 | G | A | -1.5005 | |
| 129 | L | A | 0.0000 | |
| 130 | K | A | -2.0311 | |
| 131 | L | A | 0.0000 | |
| 132 | G | A | 0.0000 | |
| 133 | I | A | 0.0000 | |
| 134 | Y | A | 0.0000 | |
| 135 | A | A | 0.0000 | |
| 136 | D | A | 0.0000 | |
| 137 | V | A | 0.0132 | |
| 138 | G | A | 0.0000 | |
| 139 | N | A | -1.9437 | |
| 140 | K | A | -2.0568 | |
| 141 | T | A | 0.0000 | |
| 142 | C | A | 0.0101 | |
| 143 | A | A | -0.3195 | |
| 144 | G | A | -1.1679 | |
| 145 | F | A | 0.0000 | |
| 146 | P | A | -1.5469 | |
| 147 | G | A | 0.0000 | |
| 148 | S | A | 0.0000 | |
| 149 | F | A | 0.0000 | |
| 150 | G | A | 0.1146 | |
| 151 | Y | A | 0.7612 | |
| 152 | Y | A | 0.6958 | |
| 153 | D | A | 0.0191 | |
| 154 | I | A | 0.2991 | |
| 155 | D | A | 0.0000 | |
| 156 | A | A | 0.0000 | |
| 157 | Q | A | -1.7088 | |
| 158 | T | A | -0.9515 | |
| 159 | F | A | 0.0000 | |
| 160 | A | A | -1.7597 | |
| 161 | D | A | -2.4847 | |
| 162 | W | A | -1.7816 | |
| 163 | G | A | -1.7950 | |
| 164 | V | A | 0.0000 | |
| 165 | D | A | -0.7232 | |
| 166 | L | A | 0.0619 | |
| 167 | L | A | 0.0000 | |
| 168 | K | A | 0.0000 | |
| 169 | F | A | 0.0000 | |
| 170 | D | A | -0.0353 | |
| 171 | G | A | 0.3029 | |
| 172 | C | A | 0.6472 | |
| 173 | Y | A | 0.7753 | |
| 174 | C | A | -0.1727 | |
| 175 | D | A | -1.7489 | |
| 176 | S | A | -0.8347 | |
| 177 | L | A | -0.4526 | |
| 178 | E | A | -2.7357 | |
| 179 | N | A | -2.5998 | |
| 180 | L | A | -1.3431 | |
| 181 | A | A | -1.6192 | |
| 182 | D | A | -2.7374 | |
| 183 | G | A | -1.5599 | |
| 184 | Y | A | 0.0000 | |
| 185 | K | A | -1.0059 | |
| 186 | H | A | -0.9864 | |
| 187 | M | A | 0.0000 | |
| 188 | S | A | -0.1744 | |
| 189 | L | A | 0.0355 | |
| 190 | A | A | 0.0000 | |
| 191 | L | A | 0.0000 | |
| 192 | N | A | -1.9977 | |
| 193 | R | A | -2.6504 | |
| 194 | T | A | -2.2312 | |
| 195 | G | A | -2.3974 | |
| 196 | R | A | -2.8714 | |
| 197 | S | A | -1.3536 | |
| 198 | I | A | 0.0000 | |
| 199 | V | A | -0.0509 | |
| 200 | Y | A | 0.0000 | |
| 201 | S | A | 0.0000 | |
| 202 | C | A | 0.0000 | |
| 203 | E | A | 0.0544 | |
| 204 | W | A | 0.7241 | |
| 205 | P | A | 0.0000 | |
| 206 | L | A | 1.3224 | |
| 207 | Y | A | 1.8828 | |
| 208 | M | A | 0.9916 | |
| 209 | W | A | 0.0000 | |
| 210 | P | A | 0.5272 | |
| 211 | F | A | 1.1095 | |
| 212 | Q | A | -1.1215 | |
| 213 | K | A | -1.9683 | |
| 214 | P | A | -1.6647 | |
| 215 | N | A | -1.9506 | |
| 216 | Y | A | 0.0000 | |
| 217 | T | A | -1.1786 | |
| 218 | E | A | -1.3373 | |
| 219 | I | A | -0.4793 | |
| 220 | R | A | -0.3381 | |
| 221 | Q | A | -0.0725 | |
| 222 | Y | A | 0.6897 | |
| 223 | C | A | 0.0000 | |
| 224 | N | A | 0.0000 | |
| 225 | H | A | -0.2352 | |
| 226 | W | A | 0.0000 | |
| 227 | R | A | 0.0000 | |
| 228 | N | A | 0.0000 | |
| 229 | F | A | -0.0581 | |
| 230 | A | A | 0.0000 | |
| 231 | D | A | -1.3956 | |
| 232 | I | A | 0.0000 | |
| 233 | D | A | -2.7580 | |
| 234 | D | A | -2.6396 | |
| 235 | S | A | -1.3617 | |
| 236 | W | A | -0.5005 | |
| 237 | K | A | -1.7747 | |
| 238 | S | A | -1.6029 | |
| 239 | I | A | 0.0000 | |
| 240 | K | A | -1.1248 | |
| 241 | S | A | -0.9887 | |
| 242 | I | A | -0.5932 | |
| 243 | L | A | -0.1649 | |
| 244 | D | A | -0.3296 | |
| 245 | W | A | 0.6793 | |
| 246 | T | A | 0.0000 | |
| 247 | S | A | 0.2512 | |
| 248 | F | A | 1.0248 | |
| 249 | N | A | 0.0000 | |
| 250 | Q | A | 0.0000 | |
| 251 | E | A | -1.8559 | |
| 252 | R | A | -1.9185 | |
| 253 | I | A | 0.0000 | |
| 254 | V | A | -1.3965 | |
| 255 | D | A | -2.0179 | |
| 256 | V | A | -1.1266 | |
| 257 | A | A | 0.0000 | |
| 258 | G | A | -0.8028 | |
| 259 | P | A | -0.5941 | |
| 260 | G | A | 0.0000 | |
| 261 | G | A | 0.0000 | |
| 262 | W | A | 0.0000 | |
| 263 | N | A | 0.0000 | |
| 264 | D | A | 0.0000 | |
| 265 | P | A | 0.0000 | |
| 266 | D | A | 0.0000 | |
| 267 | M | A | 0.0000 | |
| 268 | L | A | 0.0000 | |
| 269 | V | A | 0.0000 | |
| 270 | I | A | 0.0000 | |
| 271 | G | A | -0.5483 | |
| 272 | N | A | -0.7873 | |
| 273 | F | A | 0.7033 | |
| 274 | G | A | -0.2017 | |
| 275 | L | A | 0.0000 | |
| 276 | S | A | 0.5752 | |
| 277 | W | A | 0.4984 | |
| 278 | N | A | -0.0964 | |
| 279 | Q | A | 0.0000 | |
| 280 | Q | A | 0.0000 | |
| 281 | V | A | 0.0000 | |
| 282 | T | A | 0.0000 | |
| 283 | Q | A | 0.0000 | |
| 284 | M | A | 0.0000 | |
| 285 | A | A | 0.0000 | |
| 286 | L | A | 0.0000 | |
| 287 | W | A | 0.0000 | |
| 288 | A | A | 0.0000 | |
| 289 | I | A | 0.0000 | |
| 290 | M | A | 0.0000 | |
| 291 | A | A | 0.0000 | |
| 292 | A | A | 0.0000 | |
| 293 | P | A | -0.2295 | |
| 294 | L | A | 0.0000 | |
| 295 | F | A | 0.0000 | |
| 296 | M | A | 0.0000 | |
| 297 | S | A | 0.0000 | |
| 298 | N | A | 0.0000 | |
| 299 | D | A | -1.1702 | |
| 300 | L | A | -1.0744 | |
| 301 | R | A | -2.0412 | |
| 302 | H | A | -1.5409 | |
| 303 | I | A | -1.0806 | |
| 304 | S | A | -1.4015 | |
| 305 | P | A | -1.3610 | |
| 306 | Q | A | -1.4085 | |
| 307 | A | A | 0.0000 | |
| 308 | K | A | -1.8522 | |
| 309 | A | A | -1.0816 | |
| 310 | L | A | 0.0000 | |
| 311 | L | A | 0.0000 | |
| 312 | Q | A | -1.4676 | |
| 313 | D | A | -1.5966 | |
| 314 | K | A | -2.1713 | |
| 315 | D | A | -1.3330 | |
| 316 | V | A | 0.0000 | |
| 317 | I | A | -0.9214 | |
| 318 | A | A | -1.1525 | |
| 319 | I | A | 0.0000 | |
| 320 | N | A | -0.9350 | |
| 321 | Q | A | -1.5567 | |
| 322 | D | A | -1.2324 | |
| 323 | P | A | 0.0000 | |
| 324 | L | A | -0.4835 | |
| 325 | G | A | -0.5616 | |
| 326 | K | A | 0.0000 | |
| 327 | Q | A | -0.8349 | |
| 328 | G | A | 0.0000 | |
| 329 | Y | A | 0.0256 | |
| 330 | Q | A | -0.1369 | |
| 331 | L | A | -0.1652 | |
| 332 | R | A | -2.1963 | |
| 333 | Q | A | -2.5895 | |
| 334 | G | A | -2.3549 | |
| 335 | D | A | -2.8479 | |
| 336 | N | A | -1.9697 | |
| 337 | F | A | -1.4406 | |
| 338 | E | A | 0.0000 | |
| 339 | V | A | 0.0000 | |
| 340 | W | A | 0.0000 | |
| 341 | E | A | 0.0000 | |
| 342 | R | A | 0.0000 | |
| 343 | P | A | -0.2477 | |
| 344 | L | A | 0.0000 | |
| 345 | S | A | -0.2294 | |
| 346 | G | A | 0.3363 | |
| 347 | L | A | 1.0610 | |
| 348 | A | A | -0.2884 | |
| 349 | W | A | 0.0000 | |
| 350 | A | A | 0.0000 | |
| 351 | V | A | 0.0000 | |
| 352 | A | A | 0.0000 | |
| 353 | M | A | 0.0000 | |
| 354 | I | A | 0.0000 | |
| 355 | N | A | 0.0000 | |
| 356 | R | A | -1.5865 | |
| 357 | Q | A | -1.7626 | |
| 358 | E | A | -1.5061 | |
| 359 | I | A | 0.4877 | |
| 360 | G | A | -0.2736 | |
| 361 | G | A | -0.4898 | |
| 362 | P | A | -0.5796 | |
| 363 | R | A | -0.8472 | |
| 364 | S | A | -0.7127 | |
| 365 | Y | A | -0.5024 | |
| 366 | T | A | -0.9543 | |
| 367 | I | A | 0.0000 | |
| 368 | A | A | -0.9388 | |
| 369 | V | A | 0.0000 | |
| 370 | A | A | -0.5036 | |
| 371 | S | A | -0.9467 | |
| 372 | L | A | 0.0000 | |
| 373 | G | A | -0.7184 | |
| 374 | K | A | -1.5067 | |
| 375 | G | A | -0.3225 | |
| 376 | V | A | 1.2986 | |
| 377 | A | A | 0.3550 | |
| 378 | C | A | 0.0000 | |
| 379 | N | A | -0.2682 | |
| 380 | P | A | -0.1488 | |
| 381 | A | A | 0.0139 | |
| 382 | C | A | 0.0000 | |
| 383 | F | A | 1.2527 | |
| 384 | I | A | 0.0000 | |
| 385 | T | A | 0.0000 | |
| 386 | Q | A | -1.0394 | |
| 387 | L | A | 0.0000 | |
| 388 | L | A | 0.0000 | |
| 389 | P | A | 0.2901 | |
| 390 | V | A | 0.3885 | |
| 391 | K | A | -1.5358 | |
| 392 | R | A | -2.1747 | |
| 393 | K | A | -1.0860 | |
| 394 | L | A | 1.1230 | |
| 395 | G | A | 1.5279 | |
| 396 | F | A | 2.4127 | |
| 397 | Y | A | 0.6572 | |
| 398 | E | A | -0.9014 | |
| 399 | W | A | 0.1233 | |
| 400 | T | A | -0.3701 | |
| 401 | S | A | 0.0000 | |
| 402 | R | A | -1.9607 | |
| 403 | L | A | 0.0000 | |
| 404 | R | A | -1.6249 | |
| 405 | S | A | 0.0000 | |
| 406 | H | A | -0.8995 | |
| 407 | I | A | 0.0000 | |
| 408 | N | A | -0.7705 | |
| 409 | P | A | -0.7098 | |
| 410 | T | A | -0.9783 | |
| 411 | G | A | 0.0000 | |
| 412 | T | A | 0.0000 | |
| 413 | V | A | 0.0000 | |
| 414 | L | A | 0.0000 | |
| 415 | L | A | 0.0000 | |
| 416 | Q | A | 0.0000 | |
| 417 | L | A | 0.0000 | |
| 418 | E | A | -0.9324 | |
| 419 | N | A | 0.1770 | |
| 420 | T | A | 0.2990 | |
| 421 | M | A | 1.2441 |
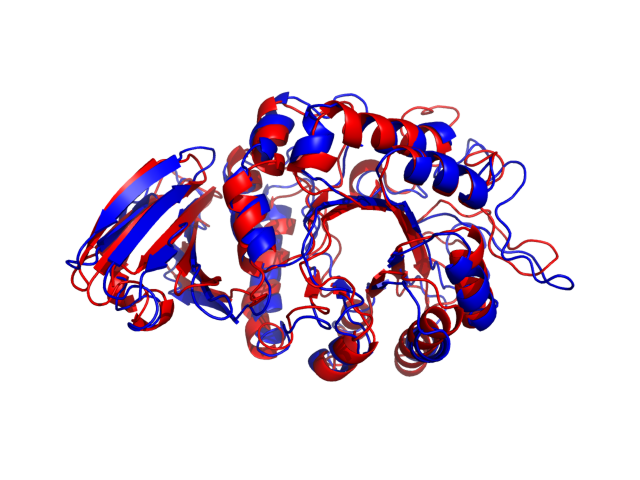
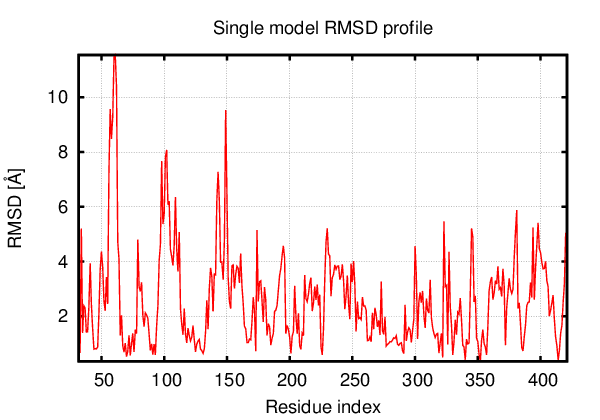
Above is the comparison between the input structure and the most aggregation prone model (predicted in the flexibility simulations, download both models in the PDB file format , download table with rmsd values ). The picture presents the most aggregation prone model (in blue) superimposed on the input structure (in red). The plot shows rmsd profile (distances between residues of the superimposed structures).
Dynamic mode uses CABS-flex simulations of protein structure fluctuations. The structure fluctuations may have impact on the size and extent of aggregation "hot-spots" on the protein surface. The dynamic mode uses the following pipeline: (1) based on the input structure, CABS-flex predicts a set of different models reflecting protein dynamics in solution; (2) for each of these models A3D score is calculated; (3) finally, the most aggregation prone model (the model with the highest A3D score, -0.6289 in this case) is selected and presented in A3D results.