In order to submit the job you need to provide:
Mark the option "Do not show my job on the queue page if you don't want the job to be visible to anyone else on the queue page.
The example of submitting the job named example_2D4F by providing the pdb code 2D4F is displayed below:
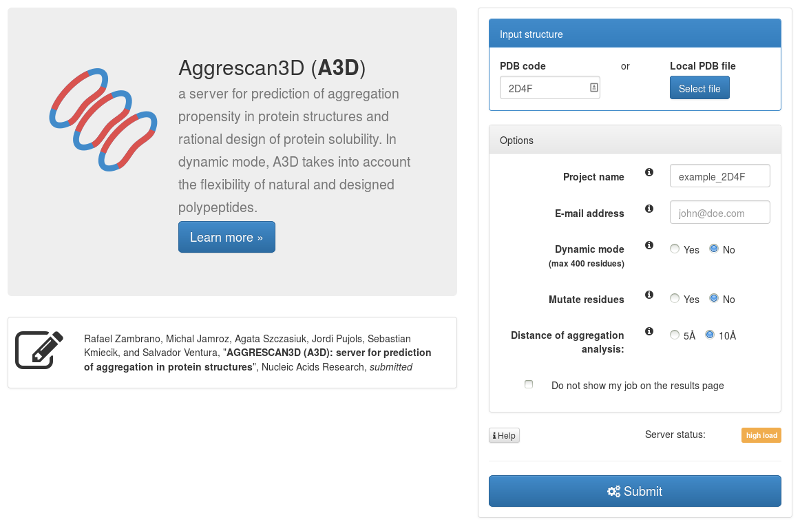
The example output of the job is provided here.
You can select to perform your predictions in Static mode (default) or Dynamic mode. In Dynamic mode A3D will take into account the flexibility of your input structure in the prediction. It should contain a single chain without breaks and no more than 400 residues.
The example output of the job, that was run in the dynamic mode, is provided here.
Dynamic mode requires input pdb files with continuous (without breaks) protein chain. pdb files with gaps in structure have to be first prepared by filling up the missing fragment.
Below is the list of example software and on-line servers that enable filling in the gaps in incomplete 3D models:
use Modeller to 'fill in' these missing residues by treating the original structure (without the missing residues) as a template, and building a comparative model using the full sequence
Availability: http://salilab.org/modeller/wiki/Missing%20residues
The server relies on the loop modeling routine in MODELLER that predicts the loop conformations by satisfaction of spatial restraints, without relying on a database of known protein structures
Availability: http://modbase.compbio.ucsf.edu/modloop/
use with "Option I: Assign additional restraints and templates to guide I-TASSER modeling." You can use a pdb file with a missing fragment as a template and provide a missing fragment sequence in an alignment file
Availability: http://zhanglab.ccmb.med.umich.edu/I-TASSER/
Users can submit a protein structure with one or more missing parts (called loops) through the Submit page and view and download the constructed loop models on the Queue page.
Availability: http://falc-loop.seoklab.org/
FREAD is a database search loop modelling algorithm. Its primary use is to fill in the gaps in incomplete 3D models of protein structures
Availability: http://opig.stats.ox.ac.uk/webapps/fread
One can use also commercially available programs for loop sampling like Prime, ICM, and Sybyl (see comparison of these programs).
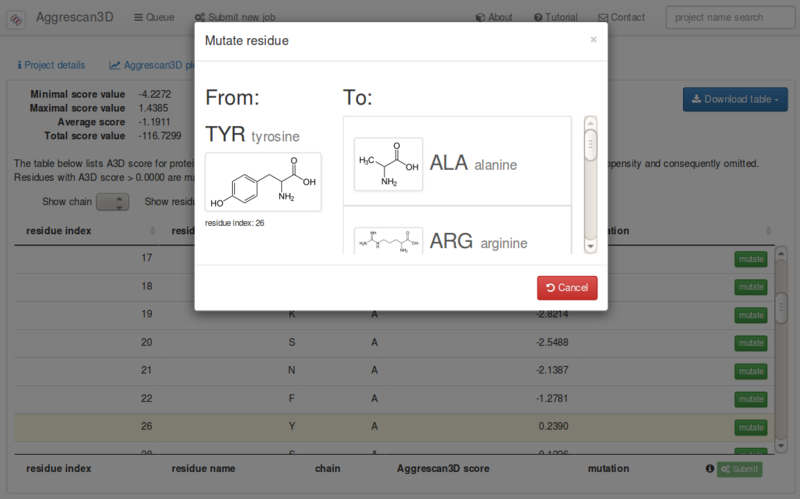
You can select to mutate one or more residues in your input structure. If you select Mutate residues upon pressing the submit button you will be directed to the “Which residues to mutate?” tab in which you can select the mutations for each desired residue and chain. The selected original and replaced residues, the position and the chain will be displayed. Save changes and submit. Mutations can also be performed previously after running a first analysis on wild type structures.
A3D can be used at two resolutions: use R = 10 Å (default) for identifying residues involved in the formation of aggregation-prone patches and R = 5 Å for a dissection of the contribution individual residues to these patches.
After clicking the Submit button, will appear info about your project details and status. If you have chosen Do not show my job on the queue page it is important to bookmark the page to check results - otherwise your job will be accessible from the queue page.
The job status information will start from the pending status, which is state when webserver waits for computational server to fetch the job input data:
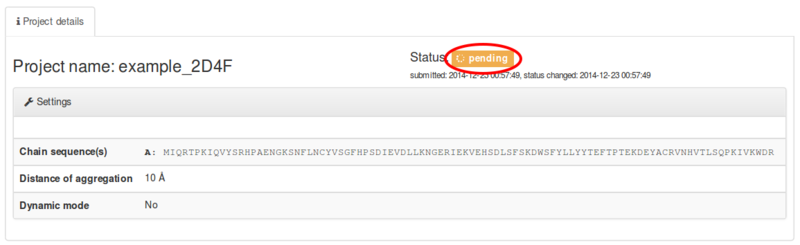
Just before computing, job status changes to running status. The jobs ends with the done status (or error status, if something went wrong admins will get email from the server about error job and will try to fix it):
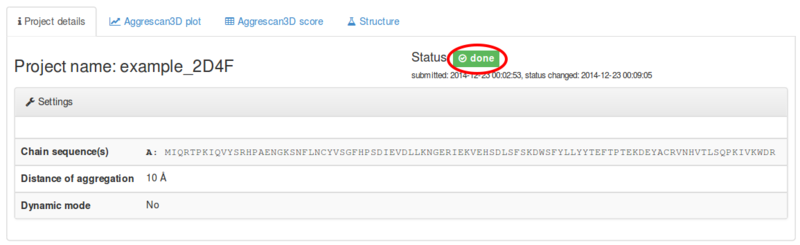
Under the Aggrescan3D score tab the following data are provided:
These values are provided for each polypeptide chain in the structure. A particular protein region in the chain can also be individually analysed.
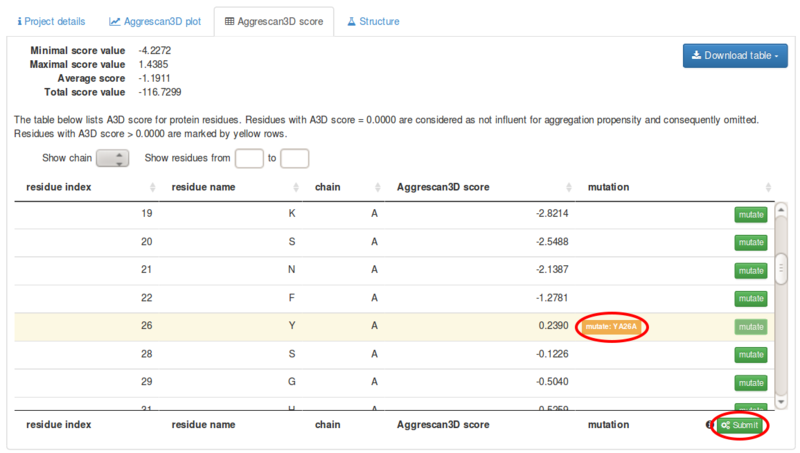
The Aggrescan3D score tab includes a scrolling list in which residue index, residue name, chain and Aggrescan3D score for all exposed residues in the structure. Residues with positive scores, i.e. aggregation-prone, are highlighted in yellow. The list can be downloaded ( button) as a csv or txt tabular data.
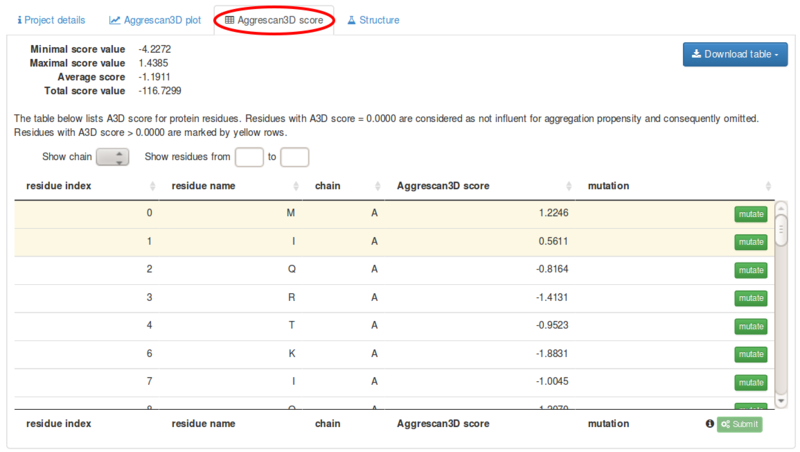
Each single residue in the list can be mutated to any of the other 19 amino acids using the button.
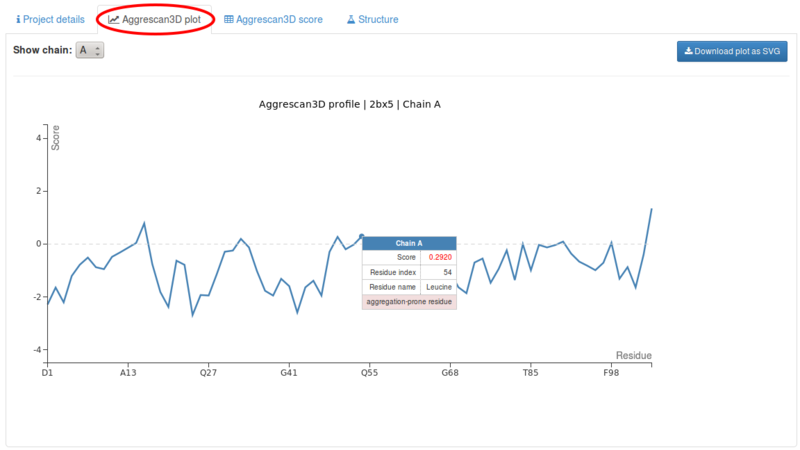
Under the Aggrescan3D plot tab the results are accessible as a line plot corresponding to the aggregation profile of the polypeptide chain. Y axis corresponds to A3D score values. Residues with positive scores in the analysed structure, therefore predicted as aggregation-prone, are indicated on top of the profile. The plot can be downloaded in svg format.
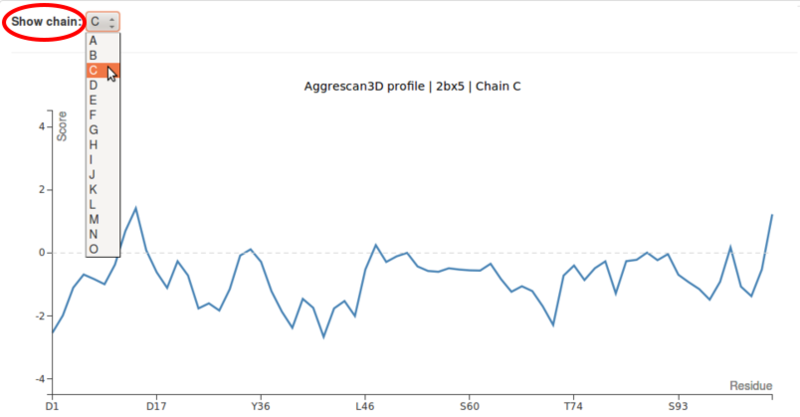
If the input structure consists of more than one chain, each chain plot can be displayed by selecting the chain signature (A, B, etc.) from the drop-down list.
Under the Structure tab a video of the protein structure is automatically generated by creating frames from pictures of the structure in different rotation states, with residues coloured according to their Aggrescan3D score. Areas with high-predicted aggregation propensity are labelled according to a red gradient and areas with high-predicted solubility according to a blue gradient. White areas are not predicted to influence aggregation (A3D score equal to zero).
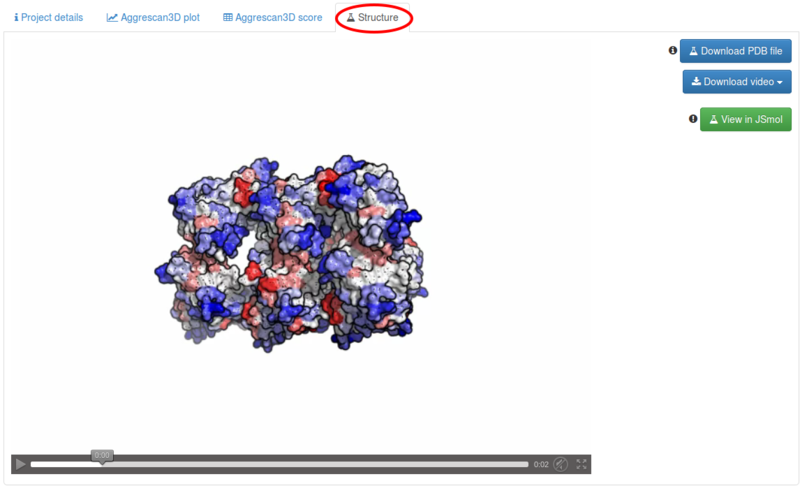
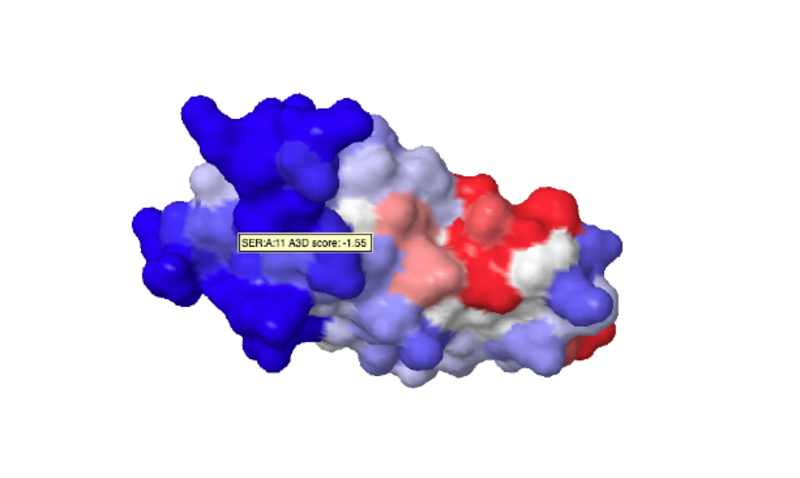
The video can be visualized and downloaded ( button) in ogv, mp4 and webm format. The correspondent pdb file can be downloaded ( button) displaying Aggrescan3D scores in the temperature factor column. Additionaly, the prediction can be visualized and inspected using jsmol ( button), as on figure below:
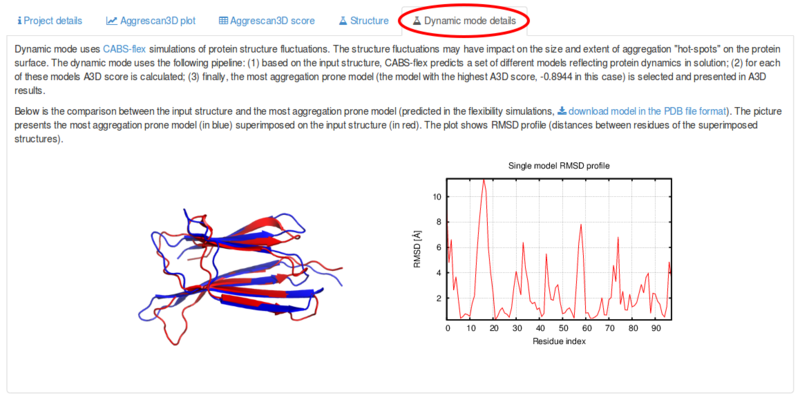
If you have selected the Dynamic mode during job submission, under the Dynamic mode details tab is provided a brief explanation of the calculation method, a structural alignment of the input structure and the most aggregation-prone model and a graphic displaying a rmsd profile (distances between residues of the two superimposed structures). Note that, the models are displayed in a random orientation. However, the most aggregation-prone model can be downloaded as a pdb file and visualized with a molecular visualization software of your choice. This model is used as the unique input structure to perform A3D predictions.
Aggrescan3D webserver may be operated throught RESTful services, using following URIs:
https://biocomp.chem.uw.edu.pl/A3D/RESTful/submit/2pcy/ submit new job (2pcy pdb entry)https://biocomp.chem.uw.edu.pl/A3D/RESTful/submit/userinput/ submit user pdb filehttps://biocomp.chem.uw.edu.pl/A3D/RESTful/submit/2pcy/chain/A/ submit new job (chain A of 2pcy pdb)
name project nameemail email (info about job will be send to this email)distance ("10" or "5") distance of aggregationdynamic ("True" or "False") use dynamic modehide ("True" or "False") if True - do not show job on the queue pagemutate - list of residues to mutate before A3D analysis. This option will be explained below.https://biocomp.chem.uw.edu.pl/A3D/RESTful/job/somejobidentifier/isdone/ check job statushttps://biocomp.chem.uw.edu.pl/A3D/RESTful/job/somejobidentifier/structure/ get output structure with A3D values in the temperature factor columnhttps://biocomp.chem.uw.edu.pl/A3D/RESTful/job/somejobidentifier/ get full information about submitted job (aggrescan3D values for individual residues, average A3D score, etc.)https://biocomp.chem.uw.edu.pl/A3D/RESTful/job/somejobidentifier/a3d_average/ Get average A3D scorehttps://biocomp.chem.uw.edu.pl/A3D/RESTful/job/somejobidentifier/a3d_min/ Get minimal A3D scorehttps://biocomp.chem.uw.edu.pl/A3D/RESTful/job/somejobidentifier/a3d_max/ Get maximal A3D scorehttps://biocomp.chem.uw.edu.pl/A3D/RESTful/job/somejobidentifier/a3d_sum/ Get total (sum) A3D scorehttps://biocomp.chem.uw.edu.pl/A3D/RESTful/job/somejobidentifier/a3d_gt0/ Get residues with A3D score > 0.0000Submit job for 2PCY protein, using cURL, with default parameters (i.e. distance of aggregation = 10Å, without mutation of residues, without distance mode, without hiding project on the queue page, without email address and with default project name): curl -i https://biocomp.chem.uw.edu.pl/A3D/RESTful/submit/2pcy/.
Submit chain B of 1hui: curl -i https://biocomp.chem.uw.edu.pl/A3D/RESTful/submit/1hui/chain/B/
User should get output like:
HTTP/1.0 200 OK
Content-Type: application/json
Content-Length: 57
Server: Werkzeug/0.9.6 Python/2.7.8
Date: Wed, 12 Nov 2014 13:52:56 GMT
{
"jobid": "a0ae2bb71feeaf0",
"status": "submitted"
}
where jobid is job identifier assigned to the just submitted job. Otherwise (for example if pdbcode doesn't exists or input data doesn't fullfill requirements), user should get output similar to:
HTTP/1.0 404 NOT FOUND Content-Type: text/html; charset=utf-8 Content-Length: 5521 Server: Werkzeug/0.9.6 Python/2.7.8 Date: Wed, 12 Nov 2014 13:56:48 GMT <!doctype html> <html lang='en'> [...]
To override default parameters, user may post additional options, i.e.: curl -i -X POST -d '{"email": "john@doe.com", "dynamic": "True", "name": "some project name"}' -H 'Content-Type: application/json' https://biocomp.chem.uw.edu.pl/A3D/RESTful/submit/2pcy/. Specified parameters will override default parameters, not specified ones would left unchanged (in this example job will be submitted with distance of aggregation = 10Å, with dynamic mode, and with specified email and project name).
The simplest Python script for submitting new job may looks like:
#!/usr/bin/env python
import requests
req = requests.post('https://biocomp.chem.uw.edu.pl/A3D/RESTful/submit/2pcy/')
print(req.status_code)
print(req.json())
Submitting with parameters (mutation) from Python script:
#!/usr/bin/env python
import requests
import json
# define which residues to mutate. If specified residues not exists within pdb
# job will not be submitted (got 404 error)
mutation_table = [{'idx': "1", 'chain': 'A', 'oldres': 'I', 'newres': 'W'},
{'idx': "2", 'chain': 'A', 'oldres': 'D', 'newres': 'W'}]
# specify additional options
options = {'dynamic': True,
'distance': 5,
'email': 'john@doe.gov',
'name': 'some project name',
'hide': True,
'mutate': mutation_table}
req = requests.post('https://biocomp.chem.uw.edu.pl/A3D/RESTful/submit/2pcy/',
data=json.dumps(options),
headers={'content-type': 'application/json'})
print(req.status_code) # get HTTP code. If 200 = OK. Otherwise = problems
print(req.json()) # get query returned data
The simplest Python script for submitting new job may looks like:
#!/usr/bin/env python
import requests
req = requests.post('https://biocomp.chem.uw.edu.pl/A3D/RESTful/submit/userinput/', files={'inputfile': open('some_file.pdb', 'rb')})
print(req.status_code)
Submitting user file with parameters (mutation) from Python script:
#!/usr/bin/env python
import requests
import json
# define which residues to mutate. If specified residues not exists within pdb
# job will not be submitted (got 404 error)
mutation_table = [{'idx': "1", 'chain': 'A', 'oldres': 'I', 'newres': 'W'},
{'idx': "2", 'chain': 'A', 'oldres': 'D', 'newres': 'W'}]
# specify additional options
options = {'dynamic': True,
'distance': 5,
'email': 'john@doe.gov',
'name': 'some project name',
'hide': True,
'mutate': mutation_table}
req = requests.post('https://biocomp.chem.uw.edu.pl/A3D/RESTful/submit/userinput/', data=options, files={'inputfile': open('some_file.pdb', 'rb')})
print(req.status_code) # get HTTP code. If 200 = OK. Otherwise = problems
By running curl -i https://biocomp.chem.uw.edu.pl/A3D/RESTful/job/somejobidentifier/isdone/ user should get:
HTTP/1.0 200 OK
Content-Type: application/json
Content-Length: 19
Server: Werkzeug/0.9.6 Python/2.7.8
Date: Wed, 12 Nov 2014 14:27:49 GMT
{
"done": false
}
if job is running/pending, or
HTTP/1.0 200 OK
Content-Type: application/json
Content-Length: 18
Server: Werkzeug/0.9.6 Python/2.7.8
Date: Wed, 12 Nov 2014 14:28:33 GMT
{
"done": true
}
if job got done/error status.
To get protein structure with A3D values in the temperature factor column and save it to out file:
curl -o out https://biocomp.chem.uw.edu.pl/A3D/RESTful/job/somejobidentifier/structure/
To get full information about submitted job, user may create Python script like:
#!/usr/bin/env python
import requests
req = requests.get('https://biocomp.chem.uw.edu.pl/A3D/RESTful/job/somejobidentifier/')
print(req.status_code)
data = req.json()
for k in data.keys():
print("key: %s: %s" % (k, data[k]))
getting output similar to:
200
key: chain_sequence: A: GSFTMPGLVDSNPAPPESQEKKPLKPCCACPETKKARDACIIEKGEEHCGHLIEAHKECMRALGFKI
key: status: done
key: mutated_residues: None
key: dynamic_mode: 0
key: mutate_mode: 0
key: last_status_change_date: 2014-11-06 12:01:04
key: project_name: b530f472611c285
key: aggregation_distance: 10
key: aggrescan3Dscore: {u'max_value': 1.664, u'total_value': -95.41120000000001, u'min_value': -4.1893, u'average_value': -1.424, u'table': [{u'res_name': u'G', u'res_idx': u'1', u'chain': u'A', u'a3d': u'-0.3367'}, ... ]}
key: submission_date: 2014-11-06 11:53:12
If user would like to get Aggrescan3D score table, she may use script like:
#!/usr/bin/env python
import requests
req = requests.get('https://biocomp.chem.uw.edu.pl/A3D/RESTful/job/somejobidentifier/')
data = req.json()
a3d = data['aggrescan3Dscore']
print("AVG A3D: %s" % (a3d['average_value'])) # data is a dictionary
print("MIN A3D: %s" % (a3d['min_value']))
for row in a3d['table'][:6]:
print(row['res_idx'], row['a3d'])
to get output like:
AVG A3D: -1.424 MIN A3D: -4.1893 (u'1', u'-0.3367') (u'2', u'0.3541') (u'3', u'1.6640') (u'4', u'0.8468') (u'5', u'1.2976') (u'6', u'0.5633')
Combine two RESTful URI to wait until job is done:
#!/usr/bin/env python
import requests
import time
server = 'https://biocomp.chem.uw.edu.pl/A3D/RESTful/job/'
jobid = "caaaffa24b4bcad"
v = False
while not v:
req = requests.get(server+jobid+'/isdone/')
v = req.json()['done']
time.sleep(60) # 1 min intervals
req = requests.get(server+jobid)
data = req.json()
a3d = data['aggrescan3Dscore']
print("AVG A3D: %s" % (a3d['average_value']))
Above script will print average A3D score as soon as job get 'done' status.
Some fun with RESTful and Bash scripting. Script below should say something as soon as job is completed:
#!/usr/bin/env bash
v="false"
while [ "$v" == "false" ]
do
v=`curl -s https://biocomp.chem.uw.edu.pl/A3D/RESTful/job/somejobidentifier/isdone/ | grep 'done' |grep -o 'false'`
sleep 6
done
mplayer -user-agent Mozilla "http://translate.google.com/translate_tts?q=Job+completed.Aggrescan+3D+is+wonderful&tl=en"