
In the Options panel there are several additional options to be chosen:
Upon selecting the mutation option and running the job the user is redirected to a new page that looks like the one below:

In order to to mutate a residue:
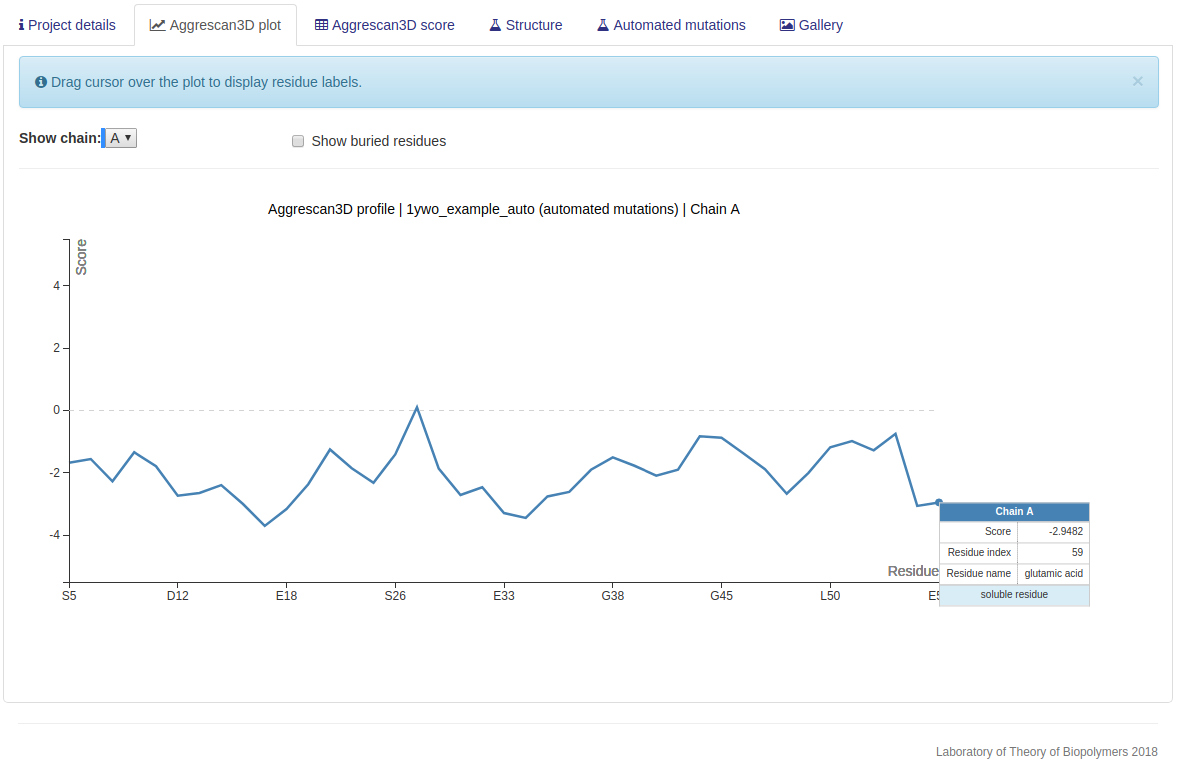
In this mode the input structure is optimized using FoldX and the most aggregation-prone residues
identified according to their A3D score. These residues are individually mutated to solubilizing charged
amino acids (arginine, aspartic acid, glutamic acid and lysine). Users can specify positions to be excluded
from the automated mutation (optional). The changes in aggregation propensity and stability are calculated for each potential point
mutant and short-listed according to these values, up to a maximum of 12 suggested changes. Only the two
most solubilizing mutations for each particular position are included in the short-list, in order to
maximize the number of residues that can be potentially engineered (up to 6).
Upon selecting this option and hitting "Run" the user is redirected to a new page:

When a job is successfully submitted the user is redirected to a summary page where all the project details are available.
This is also the place where the results are available once the calculations are finished. Depending on the chosen options
several tabs will be available.
The "Project details" tab.







Aggrescan3D 2.0 webserver may be operated through RESTful services, using following URIs:
https://biocomp.chem.uw.edu.pl/A3D2/RESTful/submit/1ywo/ submit new job (1ywo pdb entry)https://biocomp.chem.uw.edu.pl/A3D2/RESTful/submit/userinput/ submit user pdb file (details below)https://biocomp.chem.uw.edu.pl/A3D2/RESTful/submit/1ywo/chain/A/ submit new job (chain A of 1ywo pdb)
name - project name. Default: randomly generated string (jobid).email - email (info about job will be send to this email). Default: Noneemail - email (info about job will be send to this email). Default: Nonefoldx ("True" or "False") - use FoldX for stability calculations.Default: True.distance ("10" or "5") - distance of aggregation. Default: 10.dynamic ("True" or "False") - use dynamic mode. Default: Falsehide ("True" or "False") - if True - do not show job on the queue page. Default: Truemutate - list of residues to mutate before A3D analysis. This option will Illustrated in the code snippets. Default: Noneauto_mutation - selected this option to automatically enhance protein stability. Read more. Default: NoneA Python script including custom options that could be used to submit a PDB code:
#!/usr/bin/env python
import requests
import json
# define which residues to mutate. If the specified residues do not exist within the pdb file
# the server will respond with an error (code 400).
mutation_table = [{'idx': "1", 'chain': 'A', 'oldres': 'I', 'newres': 'W'},
{'idx': "2", 'chain': 'A', 'oldres': 'D', 'newres': 'W'}]
# specify additional options
options = {'dynamic': False,
'distance': 5,
'email': 'mary@sue.com',
'name': 'REST_test',
'hide': False,
'mutate': mutation_table,
'foldx': True,
'auto_mutation': "2 2"}
req = requests.post('https://biocomp.chem.uw.edu.pl/A3D2/RESTful/submit/1ywo/',
data=json.dumps(options),
headers={'content-type': 'application/json'})
print(req.status_code) # Print the HTTP code. 200 = OK. 400 = server recognises the problem, 404 = bad.
print(req.text) # Print the Response - explained below
A Python script including custom options that could be used to submit a custom PDB file:
#!/usr/bin/env python
import requests
import json
# define which residues to mutate. If the specified residues do not exist within the pdb file
# the server will respond with an error (code 400).
mutation_table = [{'idx': "1", 'chain': 'A', 'oldres': 'I', 'newres': 'W'},
{'idx': "2", 'chain': 'A', 'oldres': 'D', 'newres': 'W'}]
# specify additional options
options = {'dynamic': False,
'distance': 5,
'email': 'mary@sue.com',
'name': 'REST_test',
'hide': False,
'mutate': mutation_table,
'foldx': True,
'auto_mutation': "2 2"}
# send the request, note that the test.pdb file has to be in the same directory as this script
req = requests.post('https://biocomp.chem.uw.edu.pl/A3D2/RESTful/submit/userinput/',
files={'inputfile': ("file", open('test.pdb', 'rb')),
'json': (None, json.dumps(options), 'application/json')})
print(req.status_code) # Print the HTTP code. 200 = OK. 400 = server recognises the problem, 404 = bad.
print(req.text) # Print the Response - explained below
Other tools can be used to submit the job, as long as they are capable of POST'ing requests. Below is an example of using curl available for most unix systems for users who prefer bash scripting:
curl -i -X POST -d '{"email": "mary@sue.com", "dynamic": "True", "name": "some project name"}' -H 'Content-Type: application/json' https://biocomp.chem.uw.edu.pl/A3D2/RESTful/submit/1ywo/The responses might vary depending on the tool used, here we will focus on the Python scripts presented above
200
{
"jobid": "a0ae2bb71feeaf0",
"status": "submitted"
}
400 Auto mutation cannot be selected with dynamic nor mutate
404 <!doctype html> <html lang='en'> [...]
https://biocomp.chem.uw.edu.pl/A3D2/RESTful/job/yourjobid/status/ - Check job statushttps://biocomp.chem.uw.edu.pl/A3D2/RESTful/job/yourjobid/structure/ - Get output structure with A3D values in the temperature factor columnhttps://biocomp.chem.uw.edu.pl/A3D2/RESTful/job/yourjobid/ - Get full information about submitted job (see explained in details below)The job status can take on 5 values: pending, queue, running, error and done. See the project status in this tutorial for more explanation.
An example Python script to check the job status:
#!/usr/bin/env python
import requests
req = requests.get('https://biocomp.chem.uw.edu.pl/A3D2/RESTful/job/yourjobid/status/')
try:
data = req.json()
print(data)
except ValueError:
print("The server responded but the jobid was not found")
The likely response is:
{'status': 'done'}
An example Python script that would save the result A3D pdb file to rest_job.pdb if the job is done:
test = requests.get('https://biocomp.chem.uw.edu.pl/A3D2/RESTful/job/yourjobid/status/')
if 'done' in test.text:
req = requests.get('https://biocomp.chem.uw.edu.pl/A3D2/RESTful/job/yourjobid/structure/')
with open("rest_job.pdb", "w") as f:
f.write(req.text)
else:
print("Job not yet done")
To get full information about submitted job, user may create Python script like:
#!/usr/bin/env python
import requests
req = requests.get('https://biocomp.chem.uw.edu.pl/A3D2/RESTful/job/somejobidentifier/')
print(req.status_code)
data = req.json()
for k in data.keys():
print("key: %s: %s" % (k, data[k]))
getting output similar to:
200
key: chain_sequence: GSFTMPGLVDSNPAPPESQEKKPLKPCCACPETKKARDACIIEKGEEHCGHLIEAHKECMRALGFKI
key: status: done
key: mutated_residues: None
key: dynamic_mode: 0
key: automated_mutations: 2 3
key: stability_calculations: 1
key: mutation_energetic_effect: 0.0
key: mutation_mode: 0
key: started: 29 Jan 2019 13:19:05 GMT
key: updated: 29 Jan 2019 13:24:00 GMT
key: project_name: your_job_name
key: aggregation_distance: 10
key: aggrescan3Dscore: {u'max': 1.664,
u'sum': -95.41120000000001,
u'min': -4.1893,
u'avg': -1.424,
u'tab': [[[u'5', u'S', u'A', u'-0.3282'], [u'6', u'A', u'A', u'-0.1352'], ... ]}
key: auto_mut: {u'IE28A': [1.0209, -0.6262, -0.0538],
u'VD4P': [-0.042, -0.6319, -0.0595],
u'IR28A': [0.8208, -0.6252, -0.0528],
...}
Explaining some of the tabs:
Example script interacting with the output is presented below.
#!/usr/bin/env python
import requests
req = requests.get('https://biocomp.chem.uw.edu.pl/A3D2/RESTful/job/yourjobid/')
data = req.json() # Get the data as a json
a3d = data['aggrescan3Dscore'] # Select the aggrescan3Dscore for analysis
print("AVG A3D: %s" % (a3d['average_value'])) # print the average value - care the data is all strings at this point
print("MIN A3D: %s" % (a3d['min_value']))
for row in a3d['table'][:6]:
print(row[0], row[3])
to get output like:
AVG A3D: -1.424 MIN A3D: -4.1893 (u'1', u'-0.3367') (u'2', u'0.3541') (u'3', u'1.6640') (u'4', u'0.8468') (u'5', u'1.2976') (u'6', u'0.5633')
While we did our best to maintain backwards compatibility several performance improvements were made that might require slight modifications to the scripts meant for the previous server.