Project name: Native LECT2_Dynamic mode
Status: done
Started: 2025-05-13 04:10:53
| Chain sequence(s) |
A: GPWANICAGKSSNEIRTCDRHGCGQYSAQRSQRPHQGVDVLCSAGSTVYAPFTGMIVGQEKPYQNKNAINNGVRISGRGFCVKMFYIKPIKYKGPIKKGEKLGTLLPLQKVYPGIQSHVHIENCDSSDPTAYL
B: GPWANICAGKSSNEIRTCDRHGCGQYSAQRSQRPHQGVDVLCSAGSTVYAPFTGMIVGQEKPYQNKNAINNGVRISGRGFCVKMFYIKPIKYKGPIKKGEKLGTLLPLQKVYPGIQSHVHIENCDSSDPTAYL input PDB |
| Selected Chain(s) | A,B |
| Distance of aggregation | 10 Å |
| FoldX usage | Yes |
| pH calculations | No |
| alphaCutter usage | No |
| Dynamic mode | Yes |
| Automated mutations | No |
| Downloads | Download all the data |
| Simulation log |
[INFO] Logger: Verbosity set to: 2 - [INFO] (00:00:01)
[WARNING] runJob: Working directory already exists (possibly overwriting previous results -ow
to prevent this behavior) (00:00:01)
[INFO] runJob: Starting aggrescan3d job on: input.pdb with all chain(s) selected (00:00:01)
[INFO] runJob: Creating pdb object from: input.pdb (00:00:01)
[INFO] FoldX: Starting FoldX energy minimization (00:00:01)
[INFO] CABS: Running CABS flex simulation (00:04:49)
[INFO] Analysis: Starting Aggrescan4D on model_8.pdb (01:00:51)
[INFO] Analysis: Starting Aggrescan4D on model_6.pdb (01:00:53)
[INFO] Analysis: Starting Aggrescan4D on model_11.pdb (01:00:54)
[INFO] Analysis: Starting Aggrescan4D on model_7.pdb (01:00:56)
[INFO] Analysis: Starting Aggrescan4D on model_3.pdb (01:00:57)
[INFO] Analysis: Starting Aggrescan4D on model_5.pdb (01:00:59)
[INFO] Analysis: Starting Aggrescan4D on model_1.pdb (01:01:01)
[INFO] Analysis: Starting Aggrescan4D on model_0.pdb (01:01:02)
[INFO] Analysis: Starting Aggrescan4D on model_2.pdb (01:01:04)
[INFO] Analysis: Starting Aggrescan4D on model_4.pdb (01:01:05)
[INFO] Analysis: Starting Aggrescan4D on model_10.pdb (01:01:07)
[INFO] Analysis: Starting Aggrescan4D on model_9.pdb (01:01:08)
[INFO] Analysis: Starting Aggrescan4D on input.pdb (01:01:10)
[INFO] Analysis: Starting Aggrescan4D on folded.pdb (01:01:16)
[INFO] Main: Simulation completed successfully. (01:01:18)
|
- Minimal score value
- -3.3204
- Maximal score value
- 1.7059
- Average score
- -0.6648
- Total score value
- -176.8367
The table below lists A4D score for protein residues. Residues with A4D score > 0.0000 are marked by yellow rows.
| residue index | residue name | chain | Aggrescan4D score | mutation |
|---|---|---|---|---|
| 1 | G | A | -0.6557 | |
| 2 | P | A | -0.6043 | |
| 3 | W | A | 0.0895 | |
| 4 | A | A | 0.3167 | |
| 5 | N | A | -0.0477 | |
| 6 | I | A | 0.0000 | |
| 7 | C | A | 0.0000 | |
| 8 | A | A | 0.0000 | |
| 9 | G | A | -0.0949 | |
| 10 | K | A | 0.0000 | |
| 11 | S | A | -0.3435 | |
| 12 | S | A | -0.9449 | |
| 13 | N | A | -0.8605 | |
| 14 | E | A | -0.2189 | |
| 15 | I | A | 0.2497 | |
| 16 | R | A | -0.6241 | |
| 17 | T | A | -0.7926 | |
| 18 | C | A | 0.0000 | |
| 19 | D | A | -1.4357 | |
| 20 | R | A | 0.0000 | |
| 21 | H | A | -1.6203 | |
| 22 | G | A | -1.3624 | |
| 23 | C | A | -0.9479 | |
| 24 | G | A | -0.6172 | |
| 25 | Q | A | 0.0000 | |
| 26 | Y | A | 0.7042 | |
| 27 | S | A | -0.2829 | |
| 28 | A | A | -0.7891 | |
| 29 | Q | A | -1.9893 | |
| 30 | R | A | 0.0000 | |
| 31 | S | A | -1.7734 | |
| 32 | Q | A | -2.0293 | |
| 33 | R | A | -1.8245 | |
| 34 | P | A | -1.1194 | |
| 35 | H | A | -0.8336 | |
| 36 | Q | A | 0.0000 | |
| 37 | G | A | 0.0000 | |
| 38 | V | A | 0.0000 | |
| 39 | D | A | 0.0000 | |
| 40 | V | A | 0.0000 | |
| 41 | L | A | 0.0000 | |
| 42 | C | A | 0.0000 | |
| 43 | S | A | 0.0000 | |
| 44 | A | A | 0.0000 | |
| 45 | G | A | 0.0000 | |
| 46 | S | A | 0.0000 | |
| 47 | T | A | -0.4469 | |
| 48 | V | A | 0.0000 | |
| 49 | Y | A | -0.2064 | |
| 50 | A | A | 0.0000 | |
| 51 | P | A | 0.0000 | |
| 52 | F | A | 0.0000 | |
| 53 | T | A | -1.7094 | |
| 54 | G | A | -1.0139 | |
| 55 | M | A | -0.5781 | |
| 56 | I | A | 0.0000 | |
| 57 | V | A | -0.7259 | |
| 58 | G | A | -1.3981 | |
| 59 | Q | A | -2.0171 | |
| 60 | E | A | -2.4190 | |
| 61 | K | A | -2.5724 | |
| 62 | P | A | -1.3019 | |
| 63 | Y | A | -0.1967 | |
| 64 | Q | A | -0.8466 | |
| 65 | N | A | -1.5298 | |
| 66 | K | A | -1.0806 | |
| 67 | N | A | 0.0000 | |
| 68 | A | A | -0.6924 | |
| 69 | I | A | 0.0000 | |
| 70 | N | A | -1.8097 | |
| 71 | N | A | -1.6475 | |
| 72 | G | A | 0.0000 | |
| 73 | V | A | 0.0000 | |
| 74 | R | A | -2.2634 | |
| 75 | I | A | 0.0000 | |
| 76 | S | A | -0.7712 | |
| 77 | G | A | 0.0000 | |
| 78 | R | A | -2.0507 | |
| 79 | G | A | -1.4725 | |
| 80 | F | A | 0.0000 | |
| 81 | C | A | -0.8657 | |
| 82 | V | A | 0.0000 | |
| 83 | K | A | -1.5401 | |
| 84 | M | A | 0.0000 | |
| 85 | F | A | 0.0000 | |
| 86 | Y | A | 0.0000 | |
| 87 | I | A | 0.0000 | |
| 88 | K | A | -1.0133 | |
| 89 | P | A | 0.0459 | |
| 90 | I | A | 0.9027 | |
| 91 | K | A | -0.5690 | |
| 92 | Y | A | 0.0810 | |
| 93 | K | A | -1.1728 | |
| 94 | G | A | -0.7698 | |
| 95 | P | A | -0.8743 | |
| 96 | I | A | -1.7924 | |
| 97 | K | A | -2.6634 | |
| 98 | K | A | -3.2039 | |
| 99 | G | A | -2.1837 | |
| 100 | E | A | -2.2596 | |
| 101 | K | A | -1.8725 | |
| 102 | L | A | -0.3949 | |
| 103 | G | A | -0.2653 | |
| 104 | T | A | -0.3784 | |
| 105 | L | A | 0.0000 | |
| 106 | L | A | -0.0767 | |
| 107 | P | A | 0.0000 | |
| 108 | L | A | 0.0000 | |
| 109 | Q | A | -0.9339 | |
| 110 | K | A | -1.5989 | |
| 111 | V | A | -0.1992 | |
| 112 | Y | A | -0.4606 | |
| 113 | P | A | -0.9341 | |
| 114 | G | A | -1.1516 | |
| 115 | I | A | 0.0000 | |
| 116 | Q | A | -0.6504 | |
| 117 | S | A | 0.0000 | |
| 118 | H | A | 0.0000 | |
| 119 | V | A | 0.0000 | |
| 120 | H | A | 0.0000 | |
| 121 | I | A | 0.0000 | |
| 122 | E | A | 0.0000 | |
| 123 | N | A | -0.7943 | |
| 124 | C | A | -0.7367 | |
| 125 | D | A | -1.6271 | |
| 126 | S | A | -0.9798 | |
| 127 | S | A | -0.6694 | |
| 128 | D | A | -0.3667 | |
| 129 | P | A | -0.0594 | |
| 130 | T | A | 0.1841 | |
| 131 | A | A | 0.6781 | |
| 132 | Y | A | 1.2567 | |
| 133 | L | A | 1.7059 | |
| 1 | G | B | -0.3728 | |
| 2 | P | B | -0.1833 | |
| 3 | W | B | 0.1115 | |
| 4 | A | B | -0.3577 | |
| 5 | N | B | -1.1156 | |
| 6 | I | B | 0.0000 | |
| 7 | C | B | 0.0000 | |
| 8 | A | B | -1.0887 | |
| 9 | G | B | -1.1594 | |
| 10 | K | B | -1.2145 | |
| 11 | S | B | -1.0333 | |
| 12 | S | B | -1.0279 | |
| 13 | N | B | -0.7197 | |
| 14 | E | B | -0.8320 | |
| 15 | I | B | 0.0574 | |
| 16 | R | B | -0.8778 | |
| 17 | T | B | -1.0769 | |
| 18 | C | B | -1.4312 | |
| 19 | D | B | -2.2539 | |
| 20 | R | B | -2.7603 | |
| 21 | H | B | -2.1208 | |
| 22 | G | B | -1.7511 | |
| 23 | C | B | -1.0955 | |
| 24 | G | B | -0.6572 | |
| 25 | Q | B | -0.2887 | |
| 26 | Y | B | -0.0950 | |
| 27 | S | B | -0.5078 | |
| 28 | A | B | 0.0000 | |
| 29 | Q | B | -2.4643 | |
| 30 | R | B | -3.3204 | |
| 31 | S | B | -2.4870 | |
| 32 | Q | B | -2.6230 | |
| 33 | R | B | -2.7374 | |
| 34 | P | B | -2.0276 | |
| 35 | H | B | -2.0034 | |
| 36 | Q | B | 0.0000 | |
| 37 | G | B | 0.0000 | |
| 38 | V | B | 0.0000 | |
| 39 | D | B | 0.0000 | |
| 40 | V | B | 0.0000 | |
| 41 | L | B | 0.0000 | |
| 42 | C | B | 0.0000 | |
| 43 | S | B | 0.0000 | |
| 44 | A | B | 0.0000 | |
| 45 | G | B | -0.3678 | |
| 46 | S | B | 0.0000 | |
| 47 | T | B | 0.0000 | |
| 48 | V | B | 0.0000 | |
| 49 | Y | B | -1.0968 | |
| 50 | A | B | 0.0000 | |
| 51 | P | B | 0.0000 | |
| 52 | F | B | -0.9103 | |
| 53 | T | B | -1.4472 | |
| 54 | G | B | 0.0000 | |
| 55 | M | B | 0.1649 | |
| 56 | I | B | 0.0000 | |
| 57 | V | B | -0.4771 | |
| 58 | G | B | -0.8637 | |
| 59 | Q | B | -1.6274 | |
| 60 | E | B | -1.8489 | |
| 61 | K | B | -2.0342 | |
| 62 | P | B | -1.3633 | |
| 63 | Y | B | -1.0917 | |
| 64 | Q | B | -2.2846 | |
| 65 | N | B | -2.3619 | |
| 66 | K | B | -2.4466 | |
| 67 | N | B | -1.5380 | |
| 68 | A | B | 0.0000 | |
| 69 | I | B | 0.0000 | |
| 70 | N | B | 0.0000 | |
| 71 | N | B | 0.0000 | |
| 72 | G | B | 0.0000 | |
| 73 | V | B | 0.0000 | |
| 74 | R | B | -1.0509 | |
| 75 | I | B | 0.0000 | |
| 76 | S | B | -0.2877 | |
| 77 | G | B | -1.0510 | |
| 78 | R | B | -1.9007 | |
| 79 | G | B | -1.3338 | |
| 80 | F | B | 0.0000 | |
| 81 | C | B | -0.6637 | |
| 82 | V | B | 0.0000 | |
| 83 | K | B | -1.1069 | |
| 84 | M | B | 0.0000 | |
| 85 | F | B | -0.4416 | |
| 86 | Y | B | -0.0259 | |
| 87 | I | B | 0.0000 | |
| 88 | K | B | -0.7286 | |
| 89 | P | B | 0.0000 | |
| 90 | I | B | 0.5900 | |
| 91 | K | B | -0.9568 | |
| 92 | Y | B | -0.2308 | |
| 93 | K | B | -1.2743 | |
| 94 | G | B | 0.0000 | |
| 95 | P | B | -0.4493 | |
| 96 | I | B | 0.0000 | |
| 97 | K | B | -2.7007 | |
| 98 | K | B | -3.1007 | |
| 99 | G | B | -2.5410 | |
| 100 | E | B | -2.8002 | |
| 101 | K | B | -2.5217 | |
| 102 | L | B | -1.1664 | |
| 103 | G | B | -0.2092 | |
| 104 | T | B | -0.3255 | |
| 105 | L | B | 0.0000 | |
| 106 | L | B | 0.0695 | |
| 107 | P | B | 0.0000 | |
| 108 | L | B | 0.0000 | |
| 109 | Q | B | -0.9508 | |
| 110 | K | B | -1.1659 | |
| 111 | V | B | 0.4624 | |
| 112 | Y | B | 0.5408 | |
| 113 | P | B | -0.4671 | |
| 114 | G | B | 0.0000 | |
| 115 | I | B | 0.0000 | |
| 116 | Q | B | -0.7106 | |
| 117 | S | B | 0.0000 | |
| 118 | H | B | 0.0000 | |
| 119 | V | B | 0.0000 | |
| 120 | H | B | -0.3696 | |
| 121 | I | B | 0.0000 | |
| 122 | E | B | -1.1855 | |
| 123 | N | B | 0.0000 | |
| 124 | C | B | -1.3822 | |
| 125 | D | B | -2.6171 | |
| 126 | S | B | -1.8873 | |
| 127 | S | B | -1.3117 | |
| 128 | D | B | -1.0741 | |
| 129 | P | B | 0.0000 | |
| 130 | T | B | 0.0759 | |
| 131 | A | B | 0.3066 | |
| 132 | Y | B | 0.4713 | |
| 133 | L | B | 1.1517 |
CABS-flex predictions of flexibility of input structure
In dynamic mode, A4D analysis is performed on the set of models reflecting fluctuations of the input structure (predicted
by CABS-flex method, models are numbered from 0 to 11) and the input model.
Their A4D scores are provided below in the table.
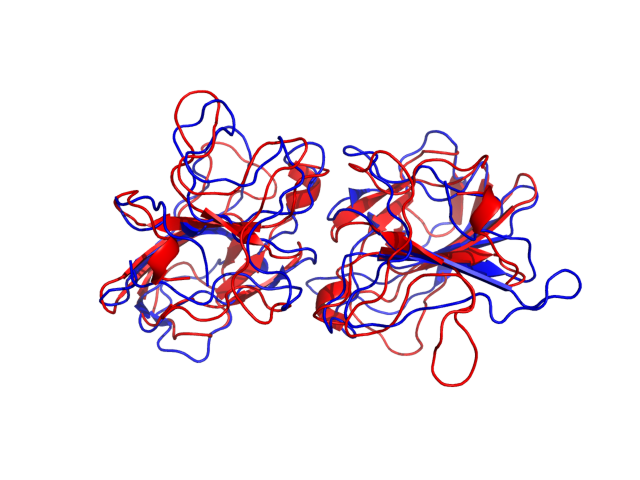
The right panel presents comparison of the most aggregation prone model (with the highest A4D score, -0.6648 in this case) with the input model
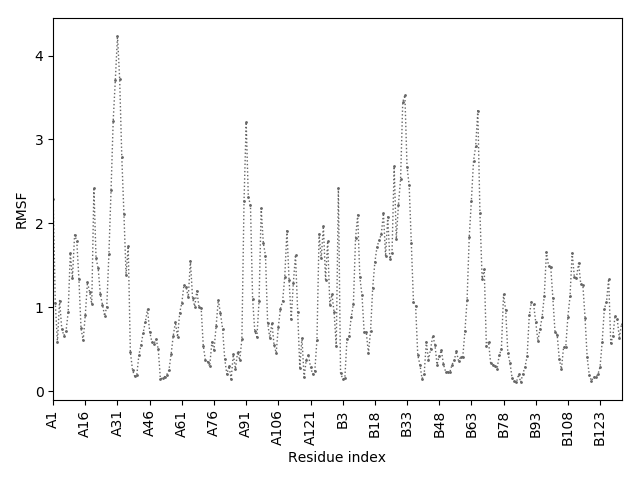
(the most aggregation prone model in blue, input in red) and RMSF plot which shows the extent of residue fluctuations in Angstroms (predicted by CABS-flex).
Model |
Average A4D Score |
|||
| model_0 | -0.6648 | View | CSV | PDB |
| model_3 | -0.6723 | View | CSV | PDB |
| model_4 | -0.7154 | View | CSV | PDB |
| model_5 | -0.7321 | View | CSV | PDB |
| model_7 | -0.7358 | View | CSV | PDB |
| model_9 | -0.7504 | View | CSV | PDB |
| model_6 | -0.7575 | View | CSV | PDB |
| CABS_average | -0.7605 | View | CSV | PDB |
| model_1 | -0.7731 | View | CSV | PDB |
| input | -0.7862 | View | CSV | PDB |
| model_11 | -0.7903 | View | CSV | PDB |
| model_2 | -0.8364 | View | CSV | PDB |
| model_8 | -0.8378 | View | CSV | PDB |
| model_10 | -0.8599 | View | CSV | PDB |
 ALA alanine
ALA alanine ARG arginine
ARG arginine ASN asparagine
ASN asparagine ASP aspartic acid
ASP aspartic acid CYS cysteine
CYS cysteine GLN glutamine
GLN glutamine GLU glutamic acid
GLU glutamic acid GLY glycine
GLY glycine HIS histidine
HIS histidine ILE isoleucine
ILE isoleucine LEU leucine
LEU leucine LYS lysine
LYS lysine MET methionine
MET methionine PHE phenylalanine
PHE phenylalanine PRO proline
PRO proline SER serine
SER serine THR threonine
THR threonine TRP tryptophan
TRP tryptophan TYR tyrosine
TYR tyrosine VAL valine
VAL valine