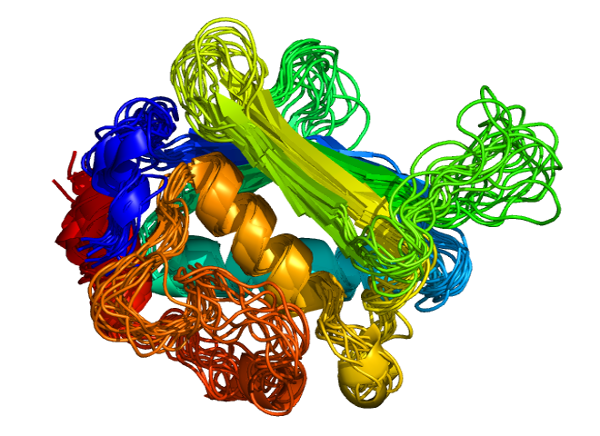
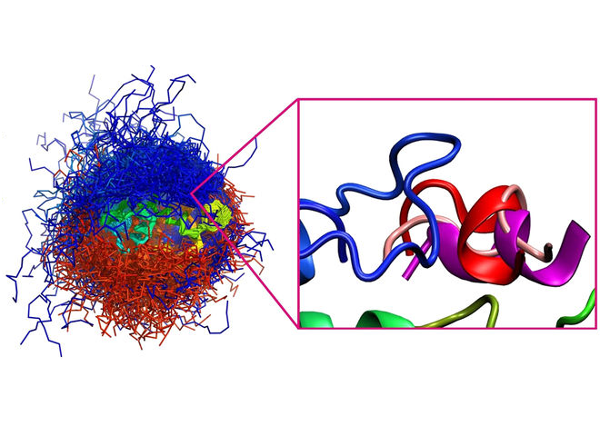
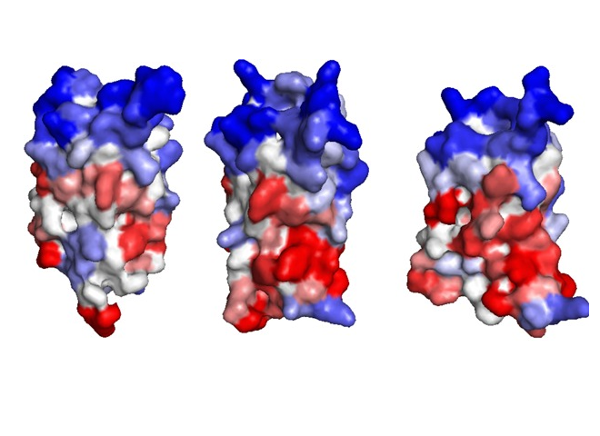
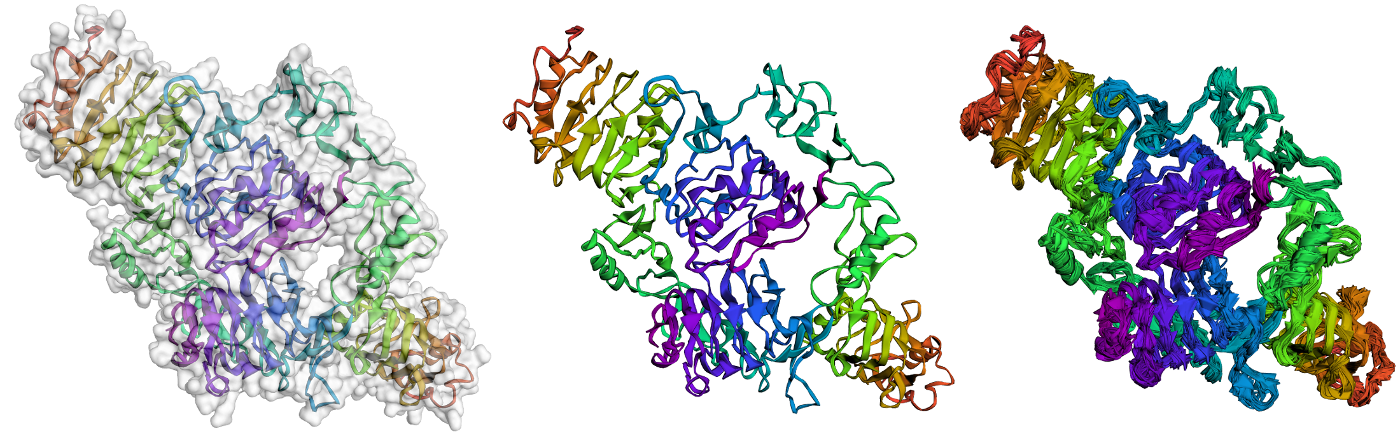
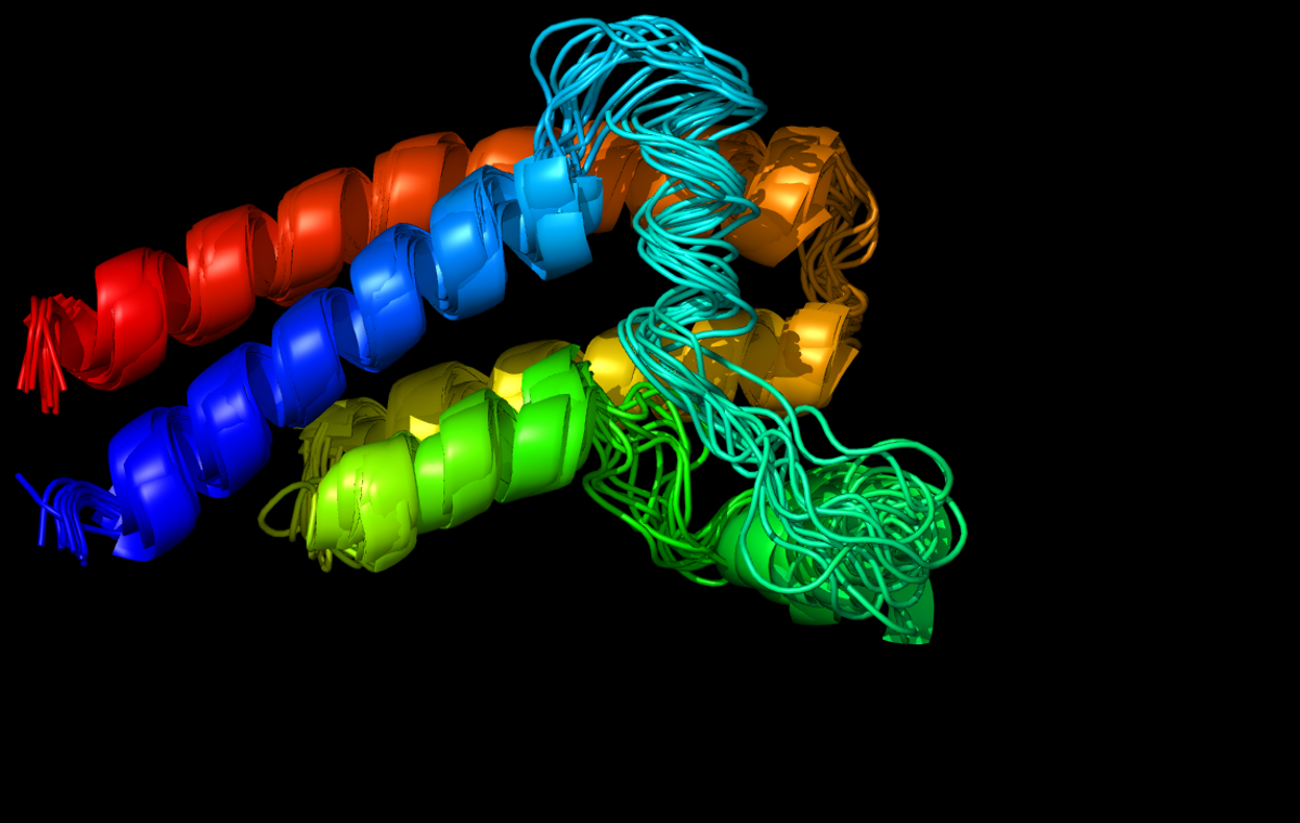
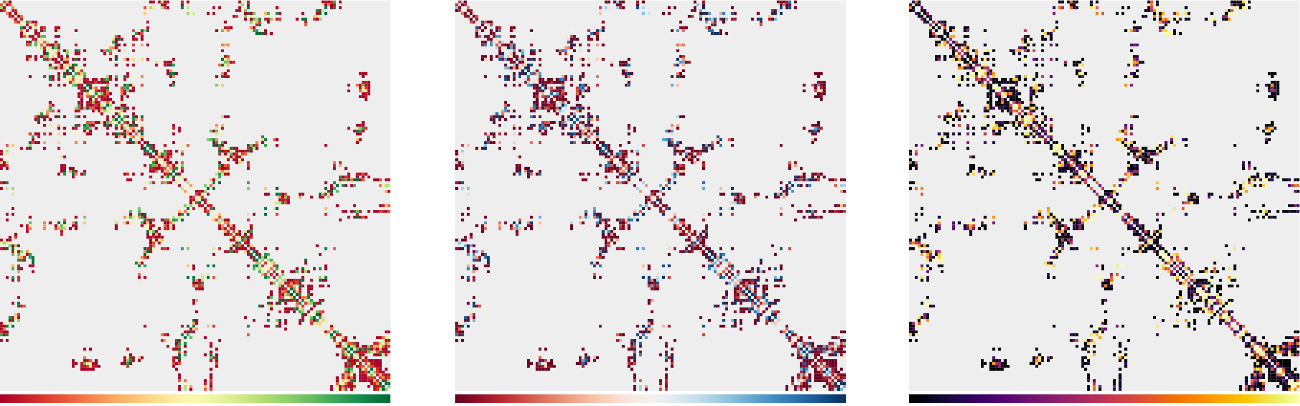
CABS-flex 2.0: a web server for fast simulations of flexibility of protein structures
Kuriata, A.*, Gierut, A. M.*, Oleniecki, T., Ciemny, M. P., Kurcinski, M., Kmiecik, S. (2018). CABS-flex 2.0: a web server for fast simulations of flexibility of protein structures. Nucleic acids research.